FITS input¶
FITS requires two types of input: Data file and Parameters file.
Data file¶
This file is expected to hold observed allele information from the system under study. FITS expects a tab-delimited textual file, with following columns:
genfor the generation of the observationallelefor the observed statefreqfor the measured frequency for that statepositionfor the position number for which the frequency data is given (optional)
Note
FITS assumes the columns to appear in the above order.
Note
The allele with the highest frequency at the first available time point will be defined as WT (w=1).
| gen | allele | freq | position |
|---|---|---|---|
| 0 | 0 | 1 | 1 |
| 0 | 1 | 0 | 1 |
| 1 | 0 | 1 | 1 |
| 1 | 1 | 0 | 1 |
| 2 | 0 | 0.99999 | 1 |
| 2 | 1 | 1e-05 | 1 |
| 3 | 0 | 0.9999899998 | 1 |
| 3 | 1 | 1.00002e-05 | 1 |
| 4 | 0 | 0.9999899998 | 1 |
| 4 | 1 | 1.00002e-05 | 1 |
| 5 | 0 | 0.9999600016 | 1 |
| 5 | 1 | 3.99984e-05 | 1 |
You can also download an example.
Note
For each generation, the sum of frequencies for the different alleles should be 1.
Note
FITS accepts allele frequencies at a given loci. Sequencing techniques tend to vary in their accuracy, so sometimes the provided allele frequencies may be inaccurate. If using inaccurate input, FITS inferences may be inaccurate as well. Specific examples include:
- Inference of fitness of highly deleterious mutations where the accuracy threshold of sequencing is worse than the mutation rate.
- Inference of mutation rate from neutral alleles when the number of generations X the mutation rate is lower than the accuracy threshold of the sequencing.
- Inference of mutation rate or fitness when very shallow sequencing is available (due to limited sampling or limited sequence coverage).
Parameters file¶
This file provides FITS with population genetics parameters information of the system under study.
Each line in this file represents a different parameter to set, where a space exists between the name of the parameter and its value: <parameter_name> <parameter value>.
Note
If you want to put comments within the parameters file, just add # at the beginning of the comments’ lines.
You can also download an example.
General parameters¶
| Parameter name | Type | Description |
|---|---|---|
| N | Integer | Size of population |
| sample_size | Integer | Size of observed population (e.g., sequenced genomes) |
| bottleneck_size | Integer | Size of the population transferred on a bottleneck event |
| bottleneck_interval* | Integer | Number of generations separating between bottleneck events (default: 0) |
| num_alleles | Integer | Number of alleles observed in all loci |
| mutation_rateX_Y | Float | Rate of mutation of allele X to allele Y. Not required if mutation rate is to be inferred |
| fitness_alleleX | Float | Fitness value assigned to allele X. Not required if fitness is to be inferred |
| logistic_growth* | Float | 1: model the population growth throughout the generations with a logistic growth model (default: 0) |
| logistic_growth_K | Float | Logistic model - upper bound |
| logistic_growth_r | Float | Logistic model - proportionality constant |
*parameter value of 0 means disabled/off; positive values mean enabled/on.
ABC parameters¶
| Parameter name | Type | Description |
|---|---|---|
| num_samples_from_prior | Integer | How many simulations to perform |
| acceptance_rate | Float | Fraction of best simulations to utilize for the inference of the parameter. |
Single simulation¶
| Parameter name | Type | Description |
|---|---|---|
| num_generations | Integer | Number of generations to simulate |
| init_freq_alleleX | Float | Initial frequency of allele X |
Fitness inference parameters¶
| Parameter name | Type | Description |
|---|---|---|
| fitness_prior | Text | One of the following:
uniform (for Uniform distribution)
log_normal (based on Bons et al. 2018)
fitness_composite
smoothed_composite (default)
See the distribution of the above priors on a (0,2) fitness here
|
| min_fitness_alleleX | Float | The minimum fitness value (inclusive) that may be assigned to allele X |
| max_fitness_alleleX | Float | The maximum fitness value (exclusive) that may be assigned to allele X |
Mutation rate inference parameters¶
X and Y are alleles defined in the data file (i.e., 0 and 1).
| Parameter name | Type | Description |
|---|---|---|
| min_log_mutation_rateX_Y | Float | Minimum (inclusive) 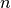 for mutation rate for mutation rate 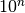 from alleleX to allele Y from alleleX to allele Y |
| max_log_mutation_rateX_Y | Float | Maximum (exclusive) 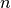 for mutation rate for mutation rate 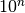 from alleleX to allele Y from alleleX to allele Y |
Population size inference parameters¶
| Parameter name | Type | Description |
|---|---|---|
| Nlog_min | Float | Minimum (inclusive) exponent 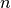 for population size for population size 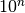 |
| Nlog_max | Float | Maximum (exclusive) exponent 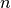 for population size for population size 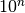 |